Biophys
The Biophysics group has focused on understanding the physical and chemical bases of protein misfolding and aggregation processes. These phenomena are of fundamental scientific and biomedical importance, as they are closely associated with the pathogenesis of serious diseases known as protein conformational disorders (PCD), including Alzheimer’s disease (AD), Parkinson’s disease (PD), and transmissible spongiform encephalopathies (TSE). Moreover, protein aggregation is gaining increasing relevance in the food industry, where it is exploited to enhance the functional properties of proteins, promoting the formation of gels or other structures useful for improving food digestibility and preservation.
Protein aggregation is driven by complex dynamics that depend on several factors such as pH, temperature, the interaction with metal ions and small molecules. Our research is mainly focused on the influence of organic protein aggregation is governed by extremely complex dynamics, influenced by various factors such as pH, temperature, and interactions with metal ions and small molecules. The research of the Tor Vergata group is primarily focused on studying the influence of organic molecules and metal ions in modulating the aggregation of proteins involved in Alzheimer’s disease (AD) and transmissible spongiform encephalopathies (TSE).
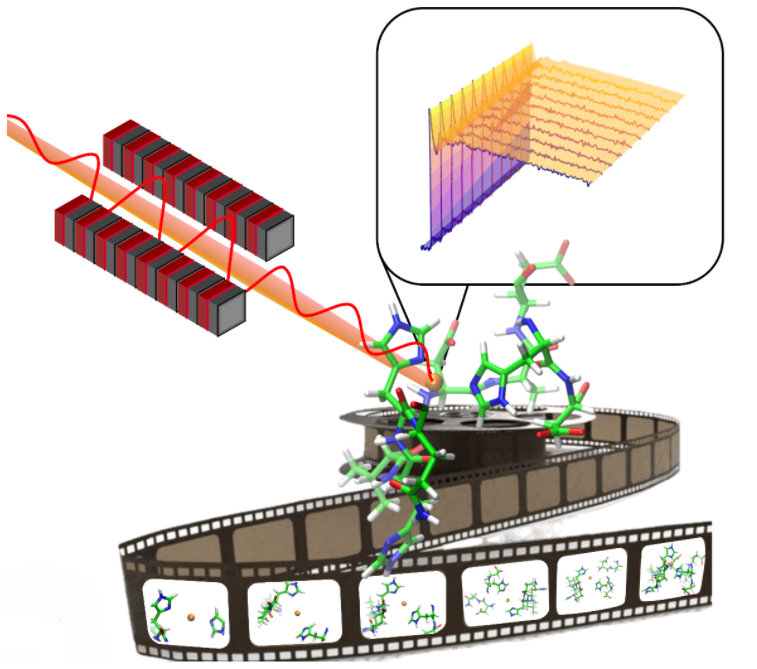
Our research strategy is based on the combination of in vitro experimental techniques, such as X-ray absorption spectroscopy (XAS), fluorescence spectroscopy, and atomic force microscopy, with modern in silico computational approaches, primarily through first-principles simulations, such as Car-Parrinello molecular dynamics. Additionally, we develop and test new numerical methods for the ab initio calculation of XAS spectra.
Useful links:
Velia Minicozzi
Francesco Stellato